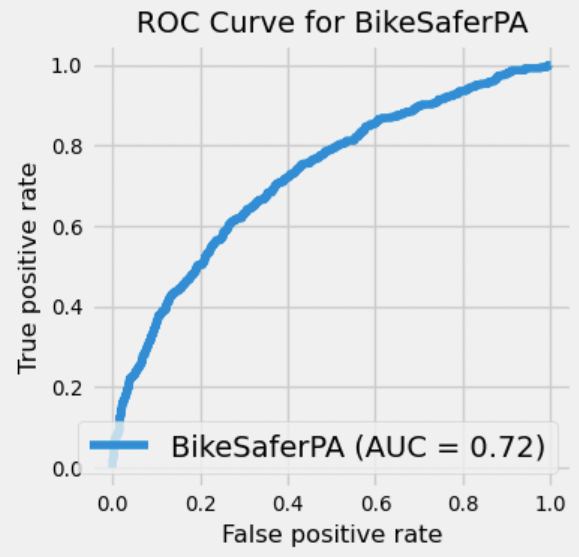
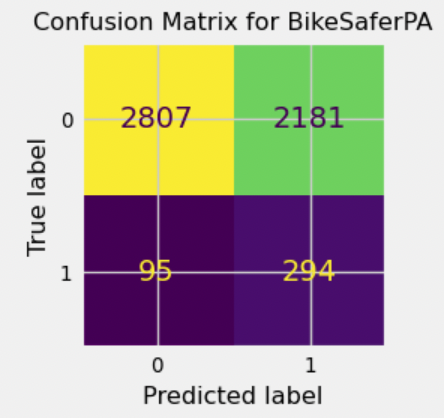
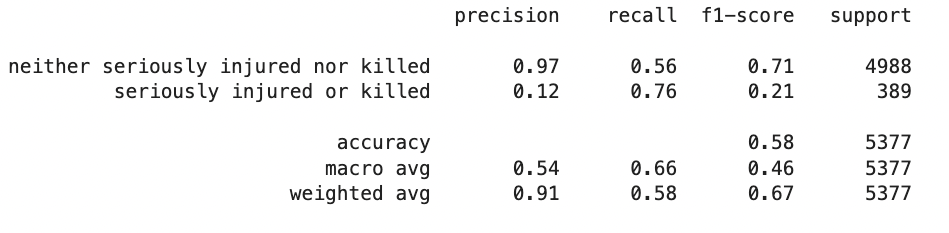
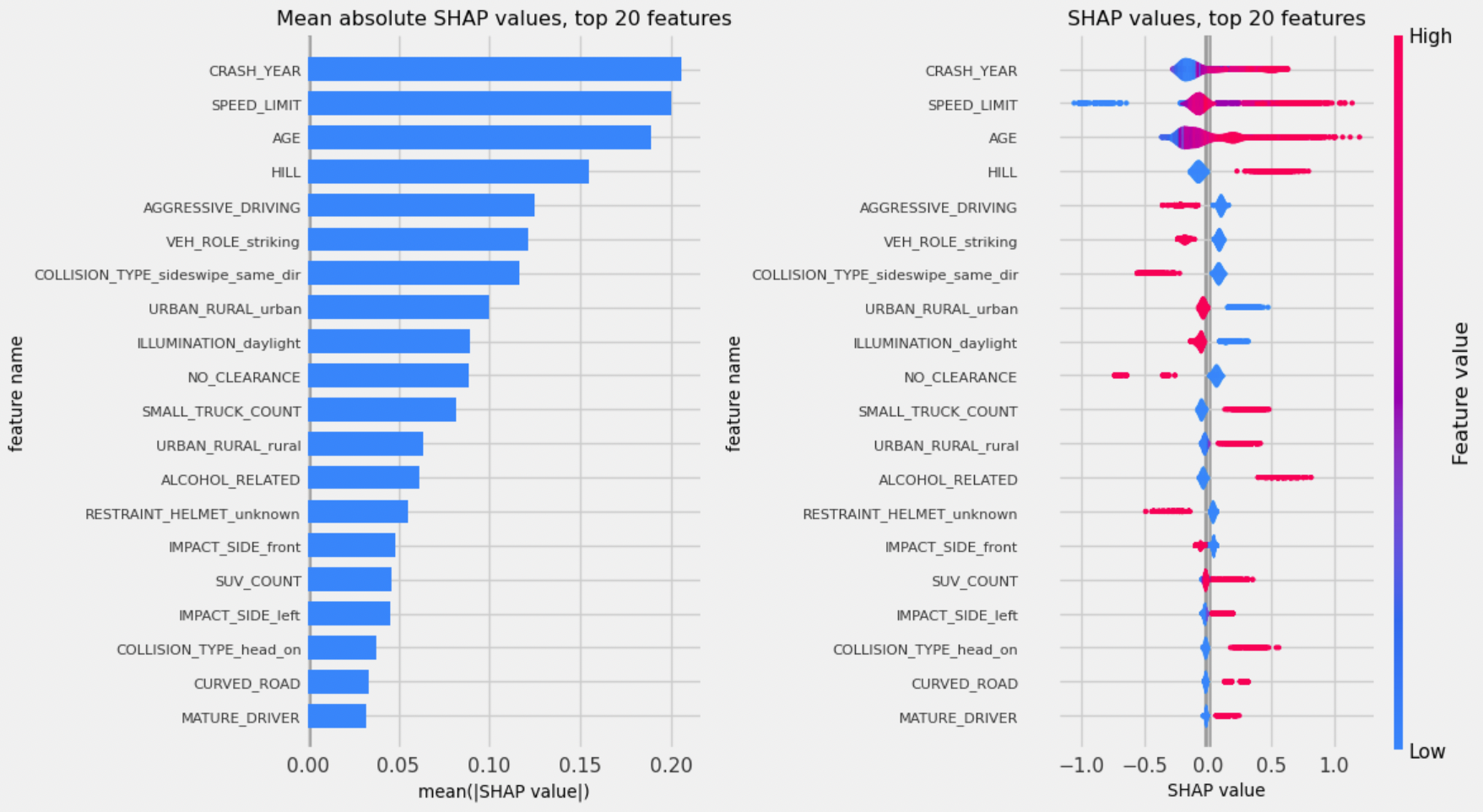
{'all': {'gb': {'learning_rate': 0.03130343103935811,
'max_depth': 2,
'min_samples_leaf': 120,
'subsample': 0.7848404943774676},
'hgb': {'l2_regularization': 11.515738930806396,
'learning_rate': 0.021772619713840338,
'max_depth': 2,
'min_samples_leaf': 143},
'lgb': {'learning_rate': 0.19669065259669294,
'max_depth': 2,
'min_child_samples': 26,
'n_estimators': 193,
'reg_alpha': 5.707852486890183,
'reg_lambda': 1.1981866373949588}},
'lr_small': {'gb': {'learning_rate': 0.03130343103935811,
'max_depth': 2,
'min_samples_leaf': 120,
'subsample': 0.7848404943774676},
'hgb': {'l2_regularization': 2.4238734679222236,
'learning_rate': 0.14182851952262965,
'max_depth': 2,
'min_samples_leaf': 140},
'lgb': {'learning_rate': 0.12656361513845585,
'max_depth': 3,
'min_child_samples': 36,
'n_estimators': 189,
'reg_alpha': 3.070386836762345,
'reg_lambda': 3.097528444873408}}}